Global Untargeted Metabolomics
Global Untargeted Metabolomics, also known as global Untargeted Metabolomics or discovery metabolomics, analyzes all small molecule metabolites simultaneously without bias.
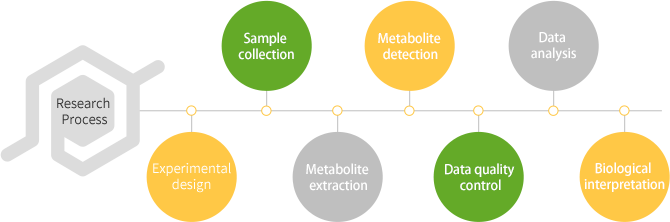
Untargeted metabolomics is used to detect and quantify the overall metabolite information of biological samples, classify samples with different treatments or physiological states, and detect through untargeted metabolomics methods to find differential metabolites from different groups, used for the study of subsequent physiological mechanisms.

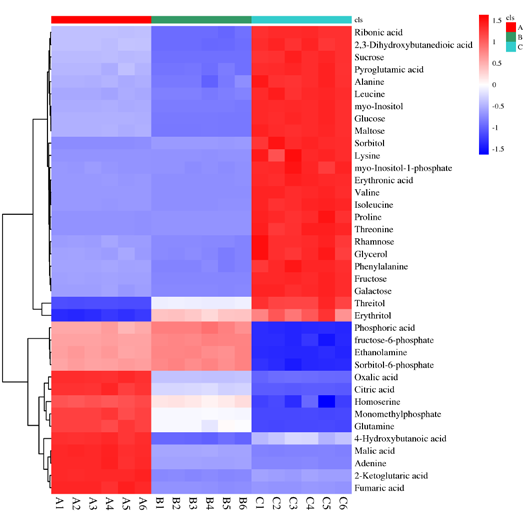 Hierarchical cluster analysis of differential metabolome
Hierarchical cluster analysis of differential metabolome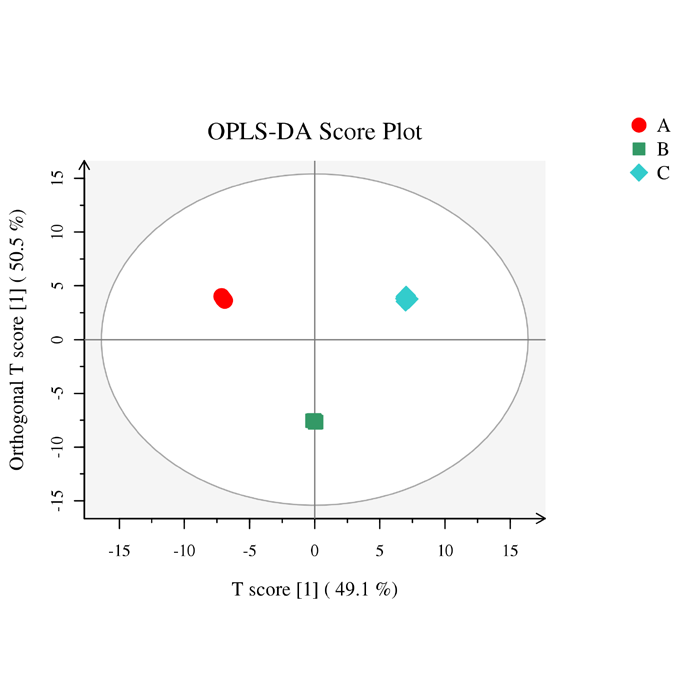 OPLS-DA_score
OPLS-DA_score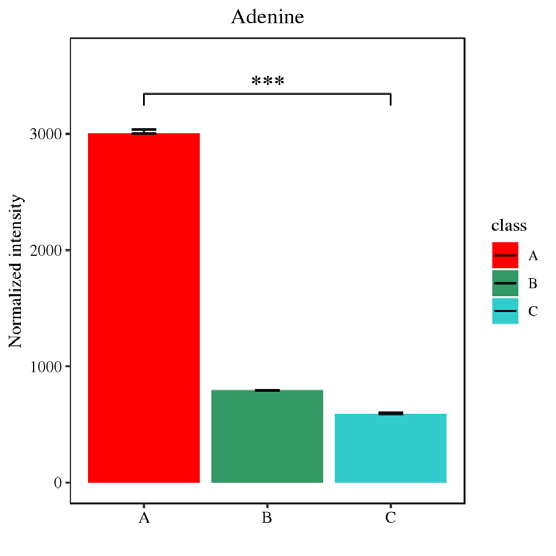 Histogram of differential metabolites
Histogram of differential metabolitesJournal: Autophagy Impact factor: 11.059 Published date: 2019 Published by: The First Affiliated Hospital of Zhejiang University School of Medicine
HGF stimulus facilitated the Warburg effect and glutaminolysis to promote biogenesis in multiple liver cancer cells.We then identified the pyruvate dehydrogenase complex (PDHC) and GLS/GLS1 as crucial substrates of HGF-activated MET kinase; MET-mediated phosphorylation inhibits PDHC activity but activates GLS to promote cancer cell metabolism and biogenesis. Moreover, we verified that Y1234/1235-dephosphorylated MET correlated with autophagy in clinical liver cancer.
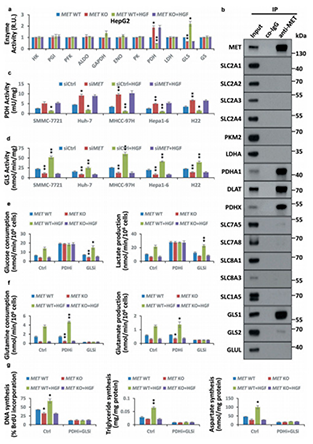 Figure 1.HGF-MET signaling promotes liver cancer metabolism and biogenesis via PDHC and GLS.
Figure 1.HGF-MET signaling promotes liver cancer metabolism and biogenesis via PDHC and GLS.
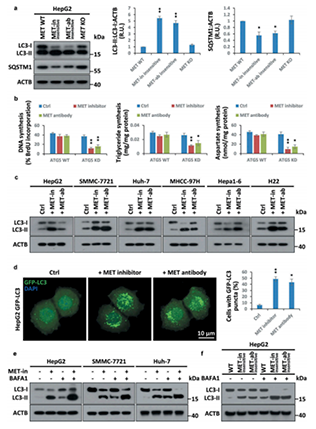 Figure 2.HGF-MET-targeted drugs induce autophagy to reset biogenesis in liver cancer
Figure 2.HGF-MET-targeted drugs induce autophagy to reset biogenesis in liver cancer
The combined application of MET inhibitors and autophagy inhibitors significantly improved the treatment efficiency of liver cancer in test tubes and mice.
Renaudin Felix,Orliaguet Lucie,Castelli Florence et al. Gout and pseudo-gout-related crystals promote GLUT1-mediated glycolysis that governs NLRP3 and interleukin-1β activation on macrophages.[J] .Ann. Rheum. Dis., 2020
 © Copyright 2015-2022 Suzhou PANOMIX Biomedical Tech Co.,Ltd
© Copyright 2015-2022 Suzhou PANOMIX Biomedical Tech Co.,Ltd