
Multi-Omics: A method for investigating the interactions between multiple substances in biological systems, including genomics, epigenomics, transcriptomics, proteomics, metabolomics, microbiomics, etc. Substances together affect the phenotype, traits, etc. of living systems. In the era of systems biology research, biological phenomena are complex and changeable, and the regulation of gene expression is complex and diverse. The conclusions of a single omics study are often not comprehensive. The combined analysis of multiple omics can realize the full spectrum analysis of proteins/transcriptions and metabolites. Explore biological issues from the two directions of "cause" and "effect", and the mutual verification effect is more obvious. It can also explain the mechanism of molecular regulation-phenotype correlation, and screen out important metabolic pathways or genes, proteins, and metabolites for experiments. analysis and research.
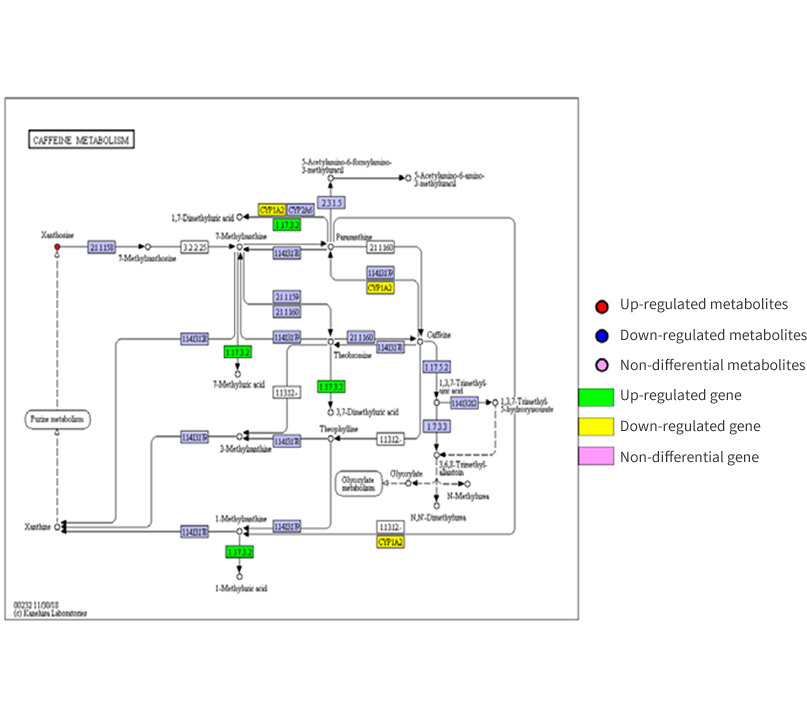 Figure 1 Integrated metabolic pathway map
Figure 1 Integrated metabolic pathway map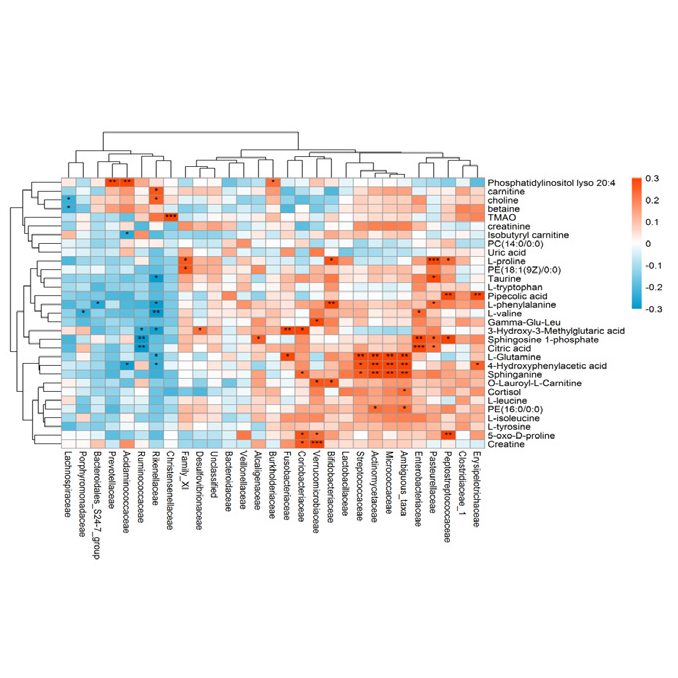 Figure 2 Correlation heatmap
Figure 2 Correlation heatmap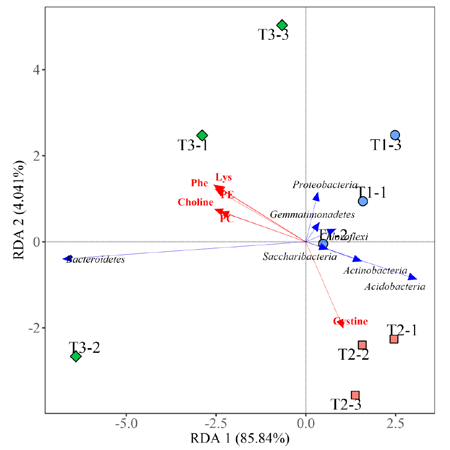 Figure 3 Redundancy analysis (Multivariate multiple linear regression relationships between response variables and explanatory variables)
Figure 3 Redundancy analysis (Multivariate multiple linear regression relationships between response variables and explanatory variables)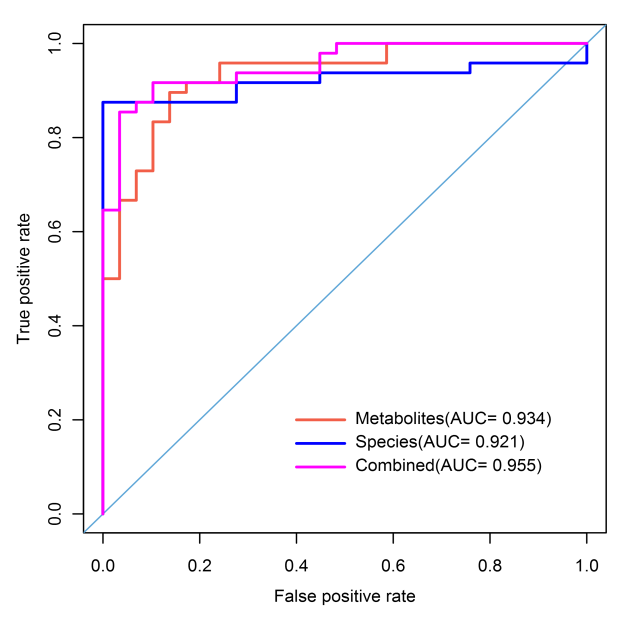 Figure 4 ROC curve (The ability of the marker to recognize and classify different groups)
Figure 4 ROC curve (The ability of the marker to recognize and classify different groups)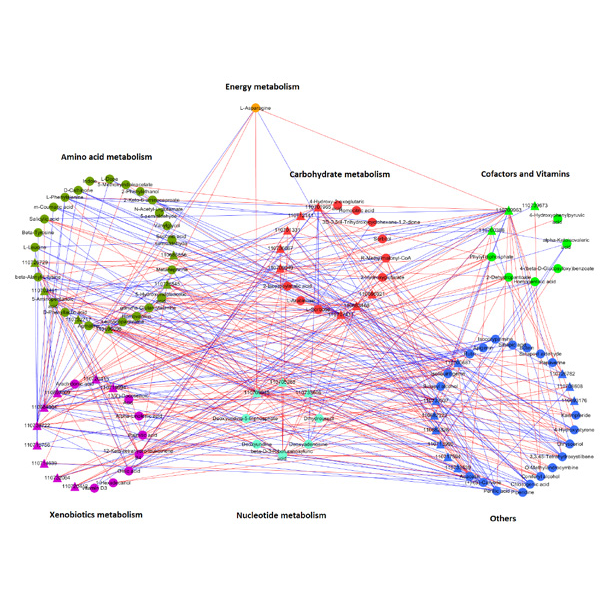 Figure 5 Network connection map based on metabolic pathways
Figure 5 Network connection map based on metabolic pathways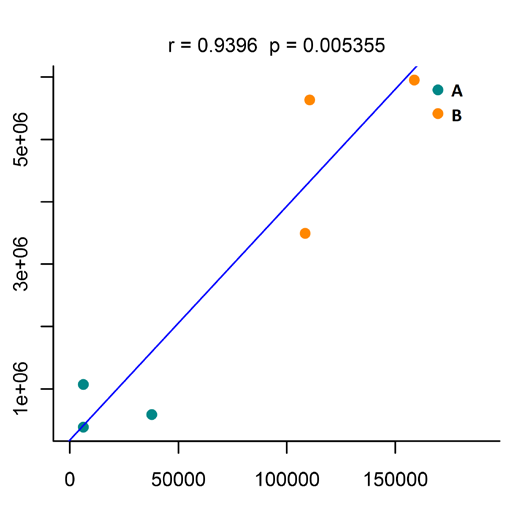 Figure 6 Scatter plot of metabolite and protein associations in a single sample
Figure 6 Scatter plot of metabolite and protein associations in a single sampleJournal: Small Impact factor: 11.459 Published date: June, 2020 Published by: Fudan University
By blocking negative immune regulatory pathways with antibodies to reactivate the immune system of body, immune checkpoint blockade therapy has achieved several clinical successes in recent years. However, due to the complexity of the tumor microenvironment and the inactivation of host immune systems, most of those immunotherapeutic strategies still have limitations in eliciting systemic antitumor responses for many cancers.Therefore, developing new approaches and strategies to effectively induce immune response and enhance antitumor response is of great significance for cancer immunotherapy.
Ferroptosis is a new cell death format identified in recent years, which can effectively inhibit tumor growth and generate mild immunogenicity, and platelet membrane-coated nanoparticles with immune evasion and tumor targeting capabilities can maximize the Ferroptosis-inducing medicine are delivered to the tumor, thereby reaching their full potential in ferroptosis therapy of tumor metastasis. The authors invented a ferroptosis-enhancing tumor immunotherapy strategy combining platelet membrane-coated sulfasalazine-loaded mesoporous magnetic nanoparticles (Fe3O4-SAS@PLT) to induce ferroptosis and PD-1 immune checkpoint blockade therapy.

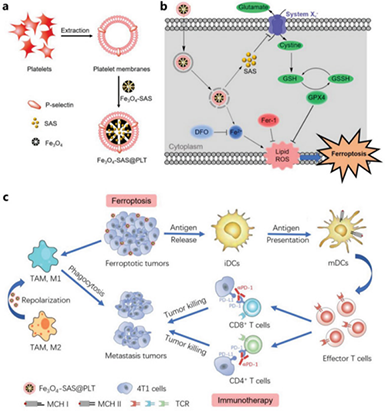 Figure1 Schematic diagram of platelet membrane-modified magnetic nanoparticles to enhance tumor immunotherapy
Figure1 Schematic diagram of platelet membrane-modified magnetic nanoparticles to enhance tumor immunotherapy 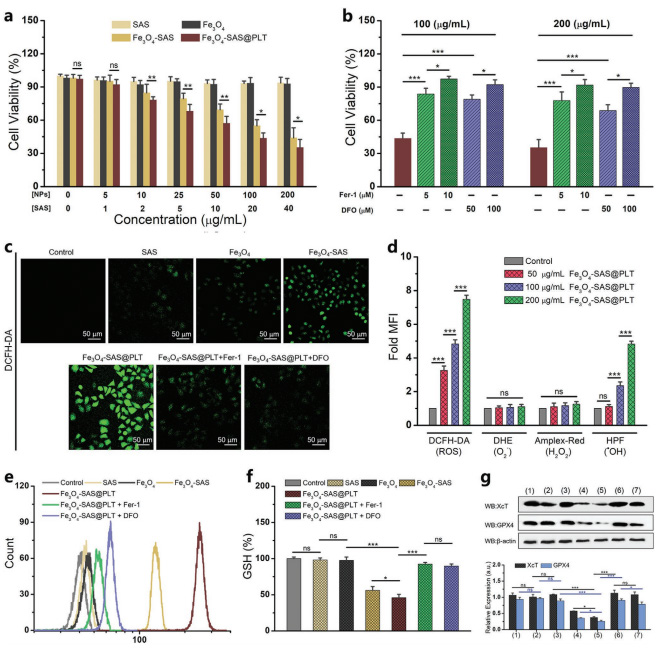 Figure 2 Study on cytotoxicity and ferroptosis mechanism of Fe3O4-SAS@PLT in vitro
Figure 2 Study on cytotoxicity and ferroptosis mechanism of Fe3O4-SAS@PLT in vitro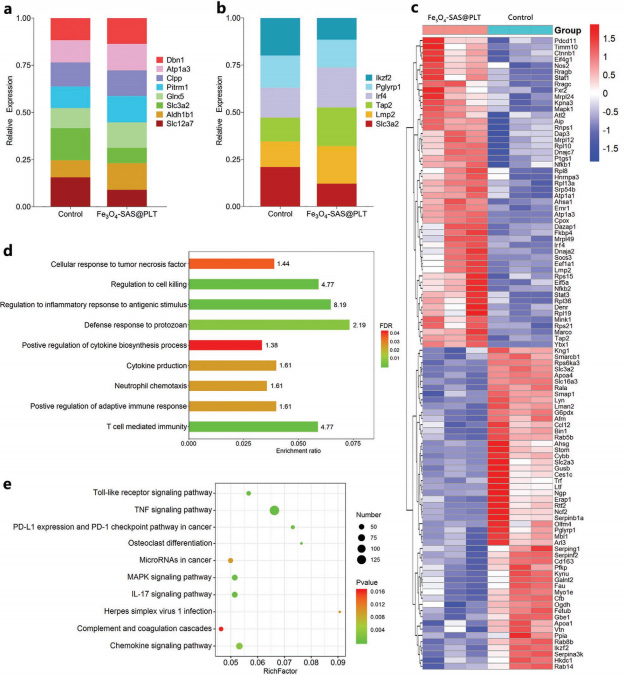 Figure 3 Mechanism of Fe3O4-SAS@PLT-mediated ferroptosis enhancing antitumor immune response
Figure 3 Mechanism of Fe3O4-SAS@PLT-mediated ferroptosis enhancing antitumor immune response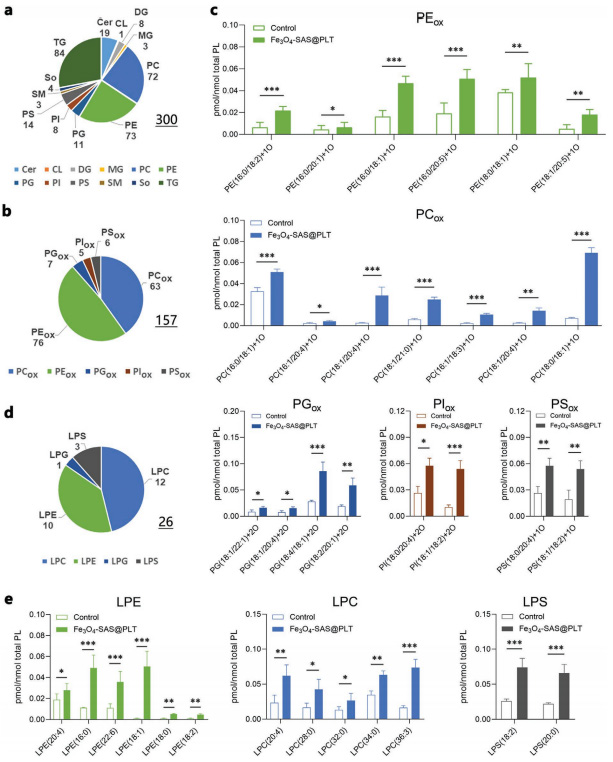 Figure 4 Fe3O4-SAS@PLT-mediated ferroptosis in vivo immune activation
Figure 4 Fe3O4-SAS@PLT-mediated ferroptosis in vivo immune activationA biomimetic magnetic nanoparticle (Fe3O4-SAS@PLT) has been developed to mediate ferroptosis and enhance tumor immunotherapy. Fe3O4-SAS@PLT nanoparticles exhibit effective immune escape and target enrichment of tumor metastasis targets. Fe3O4-SAS@PLT nanoparticles can not only induce iron-dependent ferroptosis by inhibiting the XCT pathway, but also induce effective immune response and improve the therapeutic effect of PD-1 blocking therapy in vivo. Compared with single therapy, Fe3O4-SAS@PLT mediated ferroptosis combined immunotherapy can effectively inhibit tumor growth and metastasis, and provide a new way for clinical application of synergistic immunotherapy.
Jiang Qin,Wang Kuang,Zhang Xingyu et al. Platelet Membrane-Camouflaged Magnetic Nanoparticles for FerroptosisEnhanced Cancer Immunotherapy.[J] .Small, 2020, 16: e2001704
Journal: Movement Disorders Impact factor: 8.679 Published date: 2020 Published by: University of British Columbia
Parkinson’s disease is characterized by a high burden of gastrointestinal comorbidities, especially constipation and reduced colonic transit time, and by gut microbiota alterations. The diverse metabolites produced by the microbiota are broadly relevant to host health. How microbiota composition and metabolism relate to gastrointestinal function in Parkinson’s disease is largely unknown. The objectives of the current study were to assesses associations between microbiota composition, stool consistency, constipation, and systemic microbial metabolites in Parkinson’s disease to better understand how intestinal microbes contribute to gastrointestinal disturbances commonly observed in patients.
Three hundred participants (197 Parkinson’s patients and 103 controls) were recruited for this crosssectional cohort study. Participants supplied fecal samples for microbiota sequencing (n = 300) and serum for untargeted metabolomics (n = 125). Data were collected on motor and nonmotor Parkinson’s symptoms, medications, diet, and demographics.
Significant microbiota taxonomic differences were observed in Parkinson’s patients, even when controlling for gastrointestinal function. Parkinson’s microbiota was characterized by reduced carbohydrate fermentation and butyrate synthesis capacity and increased proteolytic fermentation and production of deleterious amino acid metabolites, including p-cresol and phenylacetylglutamine. Taxonomic shifts and elevated proteolytic metabolites were strongly associated with stool consistency(a proxy for colonic transit time) and constipation among patients.

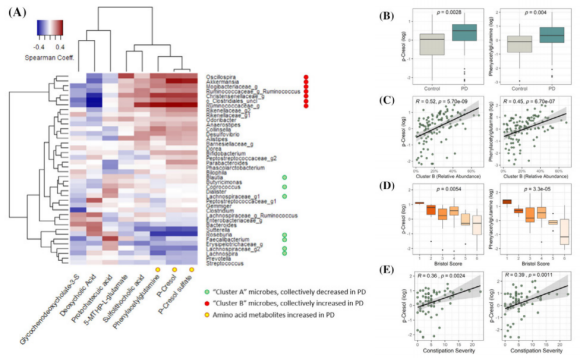
Compositional and metabolic alterations in the Parkinson’s microbiota are highly associated with gut function, suggesting plausible mechanistic links between altered bacterial metabolism and reduced gut health in this disease. The systemic detection of elevated deleterious proteolytic microbial metabolites in Parkinson’s serum suggests a mechanism whereby microbiota dysbiosis contributes to disease etiology and pathophysiology.
Cirstea Mihai S,Yu Adam C,Golz Ella et al. Microbiota Composition and Metabolism Are Associated With Gut Function in Parkinson's Disease.[J] .Mov. Disord., 2020, 35: 1208-1217
Journal: Molecular Metabolism Impact factor: 6.448 Published time: 2020 Published by: University of Rome
The metabolic influence of gut microbiota plays a pivotal role in the pathogenesis of cardiometabolic diseases. Antibiotics affect intestinal bacterial diversity and long-term usage has been identified as independent risk factor for atherosclerosis-driven events. Aim of this study was to explore the interaction between gut dysbiosis by antibiotics and metabolic pathways with impact on atherosclerosis development.
We combined oral antibiotics with different diets in an Apolipoprotein E-knockout mouse model linking gut microbiota to atherosclerotic lesion development via an integrative cross-omics approach including serum metabolomics and cecal 16S rRNA targeted metagenomic sequencing. We further investigated patients with carotid atherosclerosis compared to control subjects with comparable cardiovascular risk.
Here, we show that increased atherosclerosis by antibiotics was connected to loss of intestinal diversity and alterations of microbial metabolic functional capacity with major impact on the host serum metabolome. Pathways modulated by antibiotics and connected to atherosclerosis included diminished tryptophan and disturbed lipid metabolism. These pathways were related to reduction of certain members of Bacteroidetes and Clostridia by antibiotics in the gut. Patients with atherosclerosis presented a similar metabolic signature as the one induced by antibiotics in our mouse model.
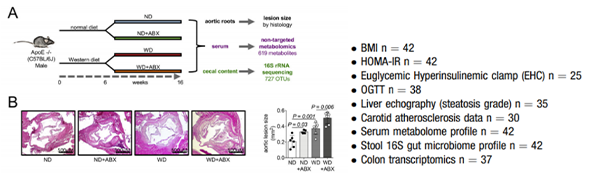 Experimental design
Experimental design
 The atherosclerosis-associated serum metabolome is associated with the reduction of certain bacteroides and clostridium in the intestine
The atherosclerosis-associated serum metabolome is associated with the reduction of certain bacteroides and clostridium in the intestine
Taken together, this work provides insights into the complex interaction between intestinal microbiota and host metabolism. Our data highlight that detrimental effects of antibiotics on the gut flora are connected to a pro-atherogenic metabolic phenotype beyond classical risk factors.
Kappel B A , Angelis L D , Heiser M , et al. Cross-omics analysis revealed gut microbiome-related metabolic pathways underlying atherosclerosis development after antibiotics treatment[J]. Molecular Metabolism, 2020
 © Copyright 2015-2022 Suzhou PANOMIX Biomedical Tech Co.,Ltd
© Copyright 2015-2022 Suzhou PANOMIX Biomedical Tech Co.,Ltd