
MobiNova-100 Single Cell Transcriptome
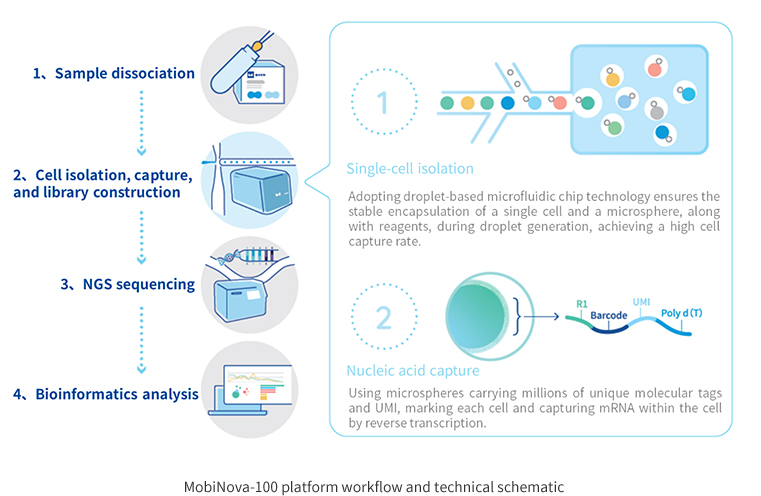
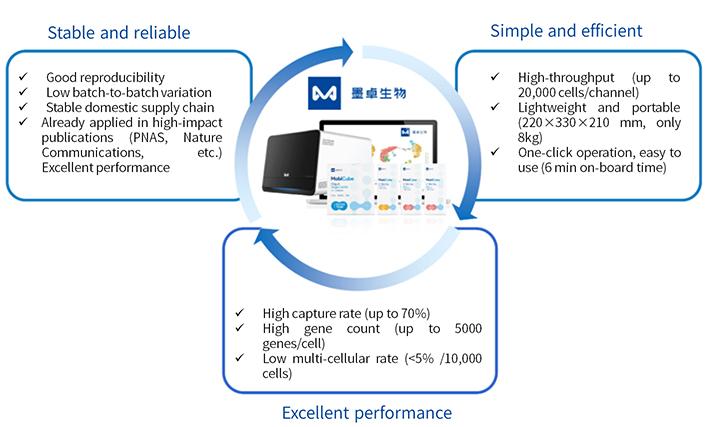
MobiNova-100® single-cell sequencing library building system is a high-quality, high-stability and cost-effective domestic single-cell sequencing platform independently developed by MobiNova, which is derived from the newly upgraded droplet microfluidic single-cell technology transformed by Professor David Weitz, a member of the Chinese Academy of Sciences IV. The MobiNova-100® uses a proprietary label and oil-in-water technology to achieve high cell capture rates by optimizing the design of the microfluidic chip and instrument control to achieve stable cell separation and co-wrapping of microspheres, reagents, and cells during droplet generation.
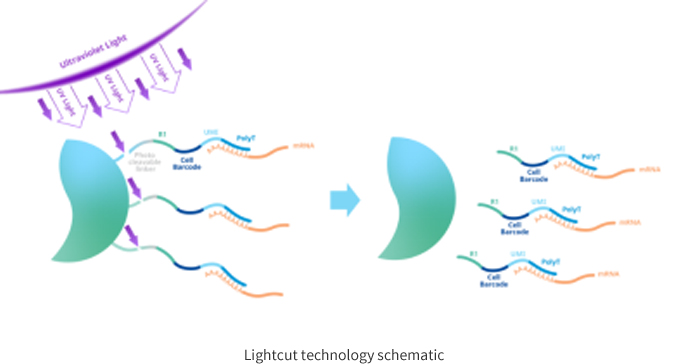
MobiNova-100 introduces the unique LightCut photo-excitation separation technology, which uses UV light irradiation to dislodge the molecular tags from the microspheres after the nucleic acid capture is completed. Compared to the 10× Genomics form of lysing microspheres, this prevents the polymeric compounds generated by lysis from entering the subsequent reaction system and thus affecting the subsequent reverse transcription reaction.
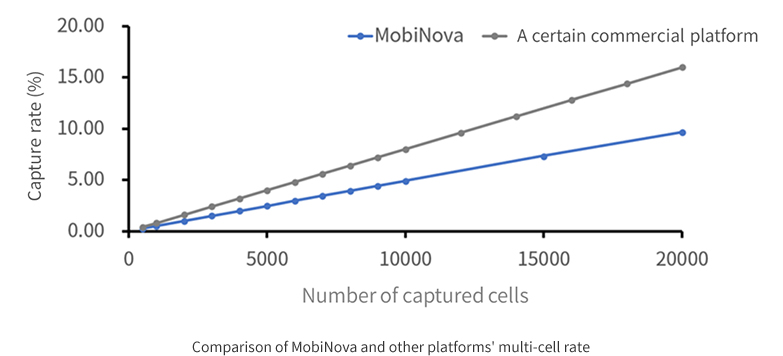
The microfluidic chip of MobiNova-100 adopts a high-speed flow channel design, which can further improve the microsphere wrapping speed and complete nucleic acid capture in 6 min. On the one hand, it can shorten the time of nucleic acid capture and reduce the degradation of mRNA molecules; on the other hand, it can make a cell match more microsphere molecules, thus reducing the rate of multicellularity and the waste of sequencing data.
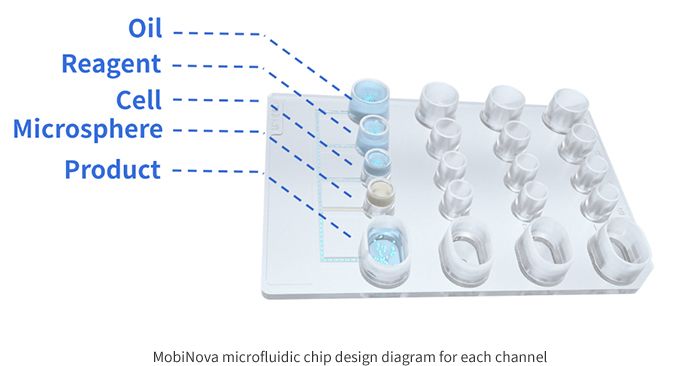
The MobiNova-100 microfluidic chip is designed with cell and reagent "partitioning" and a spike-in well to avoid stimulation of cells by reagents during the waiting time, thus affecting gene expression, i.e. avoiding the risk of cell stress and reducing system errors.
Published Journal:PNAS Impact Factor:11.1 Published:2021.12
Heterogeneity of tumor tissue is one of the current hot topics of research. The cell-to-cell heterogeneity within tumors poses a great difficulty for cancer diagnosis, treatment, and development of related drugs. Human multicellular systems, including organs, tumors, and embryos, are composed of heterogeneous and related single cells, but all originate from homogeneous and homogeneous cell populations. Understanding how heterogeneous and ordered biological systems develop from homogeneous and single cells is one of the fundamental questions in the life sciences. Current research suggests that the main sources of biological heterogeneity are genomic instability, programmed cell differentiation, accumulation of mutations, or stimulation of spatio-temporally ordered growth factor signals. Although the sources of single-cell heterogeneity by various biochemical and genetic factors have been progressively revealed, the mechanism of the origin of cellular heterogeneity caused by physical-mechanical factors in the microenvironment is not clear.
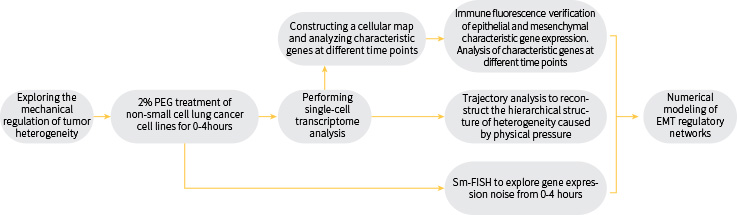
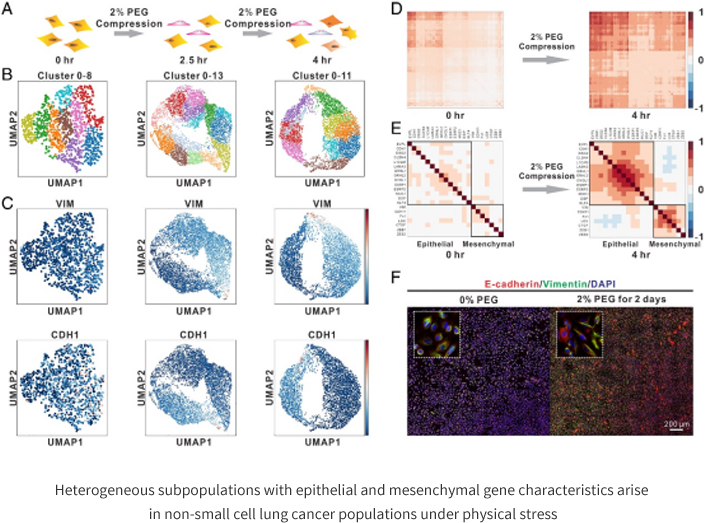
In this study, we found that a single type of mechanistic signal can induce two distinct cell phenotypes in the same homogeneous non-small cell lung cancer cells and lead to the generation of single cell heterogeneity. Further studies revealed that the mechanically induced novel cell subtypes have characteristic genes associated with cortical mesenchymal transition and characteristic genes associated with tumor stem cells. Mathematical modeling of the gene regulatory network of cortical MSC transformation revealed that the initial homogeneous non-small cell lung cancer cells were in an intermediate state, i.e., cortical/mesenchymal heterozygous state, in this regulatory network. The cells produced by force induction were located in both the "cortical" and "mesenchymal" steady states on either side of the intermediate state. Force extrusion enhances the noise (coefficient of variation) of gene expression between single cells, which causes the transcriptional profile of cells to deviate from the intermediate homeostasis in which they are located, causing cells to randomly shift to the other two homeostasis in the regulatory network, resulting in the phenomenon of bidirectional cell fate transformation.
Zhao X, Hu J, Li Y, et al. Volumetric compression develops noise-driven single-cell heterogeneity[J]. PNAS, 2021, 118(51): e2110550118.
 © Copyright 2015-2022 Suzhou PANOMIX Biomedical Tech Co.,Ltd
© Copyright 2015-2022 Suzhou PANOMIX Biomedical Tech Co.,Ltd