
Label-free Quantitative Proteomics
Journal:International Journal of Biological Sciences Impact Factor:9.2 Time:2022 Author Affiliation:ShuGuang Hospital Affiliated to Shanghai University of Chinese Traditional Medicine
Nonalcoholic fatty liver disease (NAFLD), a spectrum of liver disease characterized by fat accumulation, affects more than 25% of the global population.The molecular mechanisms of NAFLD remain incompletely understood. Lipids produced by de novo lipogenesis have been considered to be the fundamental step in the development of NAFLD and are caused by the imbalance between triglyceride (TG) synthesis and degradation. Previous studies have revealed several protein subtypes that can regulate the expression or degradation of these enzymes, such as transcription factors (TFs), RNA binding proteins, and ubiquitin ligases.
High fat chow-fed NAFLD mice were used to identify differentially expressed hepatic proteins associated with NAFLD using a label-free quantitative proteomics strategy, and key transcription factors were screened by functional analysis to uncover the physiological mechanisms involved in the regulation of NAFLD.
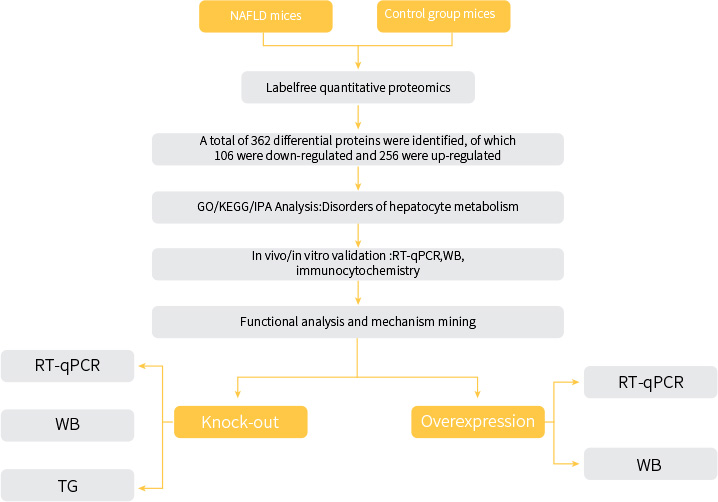
Fig.1 Methods
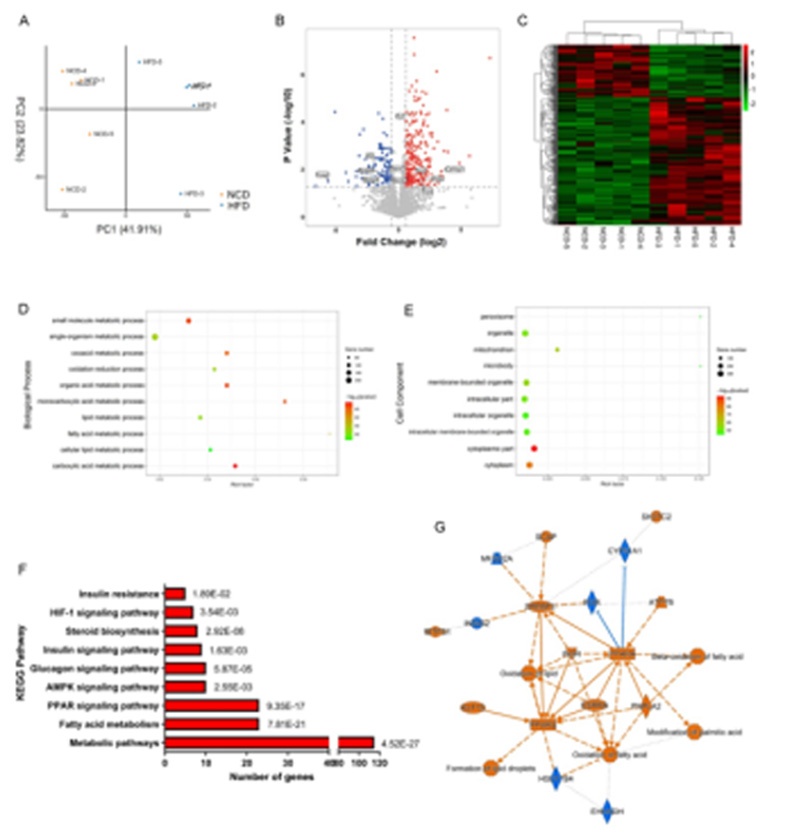
Fig.2 label-free differential protein identification, GO/KEGG enrichment analysis and IPA analysis were performed
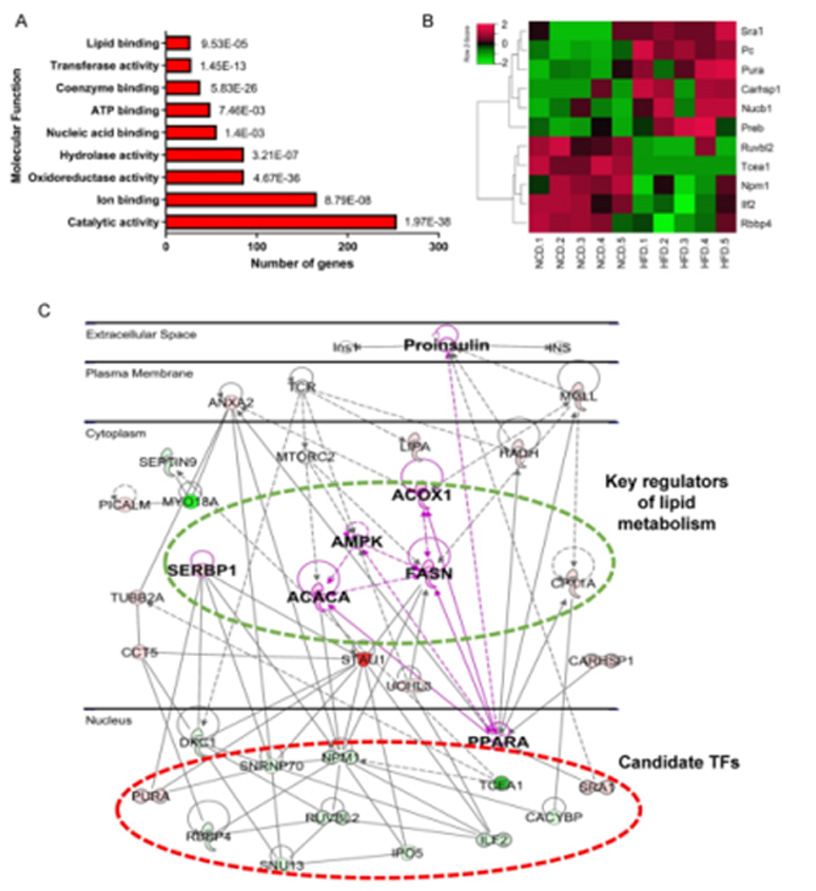
Fig.3 Bioinformatics analysis of DEPs revealed metabolic dysfunction related TFs
In this study, a comprehensive analysis of protein expression patterns and biological process perturbations involved in the development of NAFLD by quantitative proteomics identified three novel NAFLD-related TFs: Tcea1, Rbbp4 and ILF2, which were validated in NAFLD model mice and clinical liver samples. Follow-up functional analysis showed that down-regulation of all three transcription factors significantly promoted lipid accumulation in AML12 hepatocytes by regulating the expression of fatty acid synthesis or β-oxidation-related genes. In contrast, overexpression of Tcea1, Rbbp4 and ILF2 ameliorated hepatocyte steatosis. These findings suggest new lipid metabolism-related TFs that may play a potential role in the prevention of NAFLD.
Zhi S, Congcong Z, Zhiling G, et al. Quantitative proteomics of HFD-induced fatty liver uncovers novel transcription factors of lipid metabolism[J]. Int J Biol Sci, 2022.
 © Copyright 2015-2022 Suzhou PANOMIX Biomedical Tech Co.,Ltd
© Copyright 2015-2022 Suzhou PANOMIX Biomedical Tech Co.,Ltd