
Diversity of Microbial
Microbial diversity research refers to the method of detecting the microbial composition and abundance of samples by methods such as high-throughput sequencing. Microbial diversity sequencing, also known as amplicon sequencing, uses high-throughput sequencing to sequence specific regions of prokaryotic 16S rDNA or eukaryotic 18S rDNA/ITS/functional genes. Compared with traditional methods, there is no need for pure culture, which greatly reduces the cost, cycle and species richness of research.
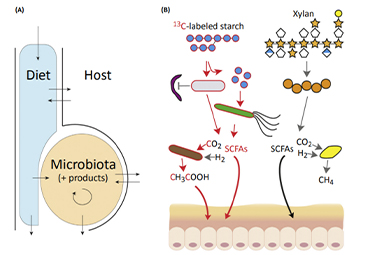
Journal: Gastroenterology Impact factor: 19.233 Published date: 2019 Published by: National University of Ireland
Irritable bowel syndrome (IBS) is a heterogeneous disorder but diagnoses and determination of sub-types are made based on symptoms. We profiled fecal microbiomes of patients with and without irritable bowel syndrome (IBS) to identify biomarkers of this disorder.
We collected fecal and urine samples from 80 patients with IBS (Rome IV criteria; 16–70 years old) and 65 matched individuals without IBS (controls), along with anthropometric, medical, and dietary information. Shotgun and 16S rRNA amplicon sequencing were performed on feces, whereas urine and fecal metabolites were analyzed by gas chromatography and liquid chromatography mass spectrometry.Bile acid malabsorption (BAM) was identified in patients with diarrhea by retention of radiolabelled selenium-75 homocholic acid taurine.
Patients with IBS had significant differences in network connections between diet and fecal microbiomes compared with controls; these were accompanied by differences in fecal metabolomes. We did not find significant differences in fecal microbiota composition among patients with different IBS symptom subtypes. Fecal metabolome profiles could discriminate patients with IBS from controls. Urine metabolomes also differed significantly between patients with IBS and controls, but most discriminatory metabolites were related to diet or medications. Fecal metabolomes but not microbiomes could distinguish patients with IBS with vs those without BAM.
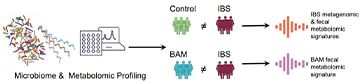 Figure 1 Graphic abstract
Figure 1 Graphic abstract
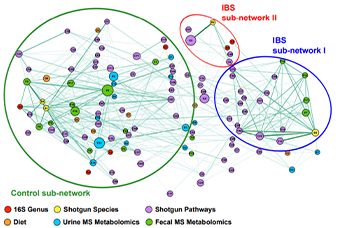 Figure 2 Visual analysis based on multi-dimensional data
Figure 2 Visual analysis based on multi-dimensional data
Despite the heterogeneity of IBS, patients have significant differences in urine and fecal metabolomes and fecal microbiome from controls, independent of symptom-based subtypes of IBS. Fecal metabolome analysis can be used to distinguish patients with IBS with vs without BAM. These findings might be used to develop microbe-based treatments for these disorders.
I. Jeffery et al., Differences in Fecal Microbiomes and Metabolomes of People With vs Without Irritable Bowel Syndrome and Bile Acid Malabsorption. Gastroenterology. 2019 Dec 13
 © Copyright 2015-2022 Suzhou PANOMIX Biomedical Tech Co.,Ltd
© Copyright 2015-2022 Suzhou PANOMIX Biomedical Tech Co.,Ltd